Recent Publications
2026
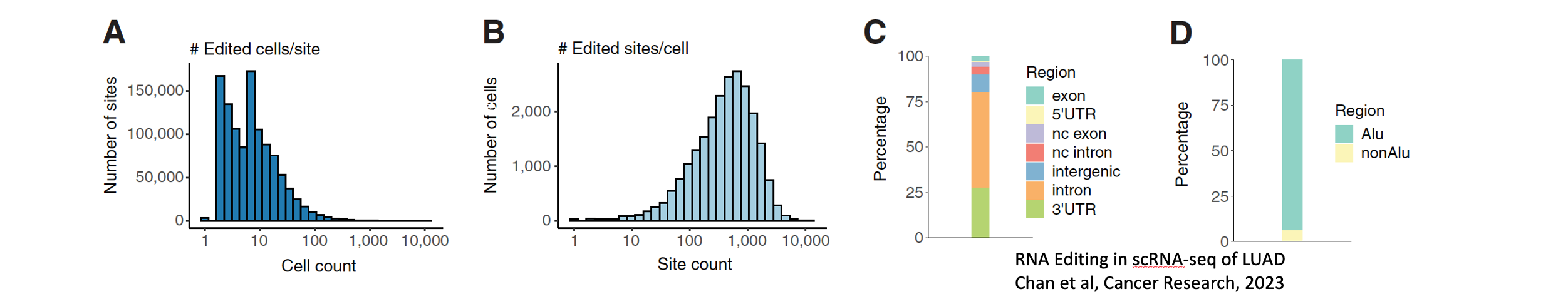
Wang, Siqi; Chen, Chuyun; Xiao, Xinshu; Wang, Zefeng
Genetic variation shapes human mRNA translation and disease risk Unpublished
bioRxiv, 2026.
@unpublished{Wang2026,
title = {Genetic variation shapes human mRNA translation and disease risk},
author = {Siqi Wang and Chuyun Chen and Xinshu Xiao and Zefeng Wang},
url = {http://biorxiv.org/lookup/doi/10.64898/2026.02.10.705206},
doi = {10.64898/2026.02.10.705206},
year = {2026},
date = {2026-02-12},
publisher = {openRxiv},
abstract = {Abstract
Genetic variation can influence protein abundance through translation, yet this regulatory layer remains poorly defined. We developed a deep learning approach to systematically map the effects of single-nucleotide variant (SNV) on translation efficiency (TE) across the human genome. In lymphoblastoid cells, >90,000 variants substantially altered TE, with strong positional and sequence-context biases. Importantly, the missense variants, traditionally considered only for their effect on protein recoding, were also found to reshape translation efficiency, with proline substitutions consistently reducing TE in a length-dependent manner. Extending our analysis to eight additional cell types revealed a two-layer architecture of translational regulation: 5’UTR variants produced highly concordant effects across cell types, whereas synonymous and missense variants in coding region exhibited cell type-specific outcomes, suggesting context-dependent translation regulation. TE-altering variants were enriched among GWAS loci and linked to cancer, immune, cardiometabolic, and neurological traits, positioning translation as a key mediator of genetic effects on disease. },
howpublished = {bioRxiv},
keywords = {},
pubstate = {published},
tppubtype = {unpublished}
}
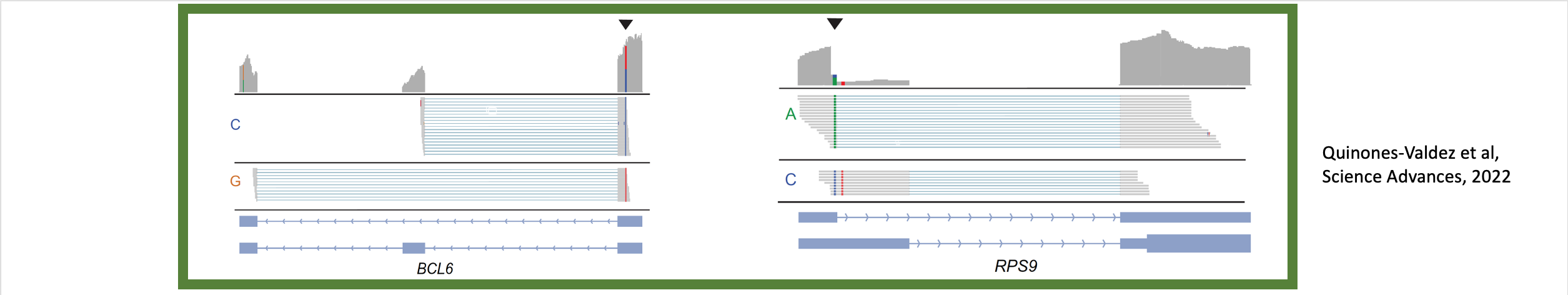
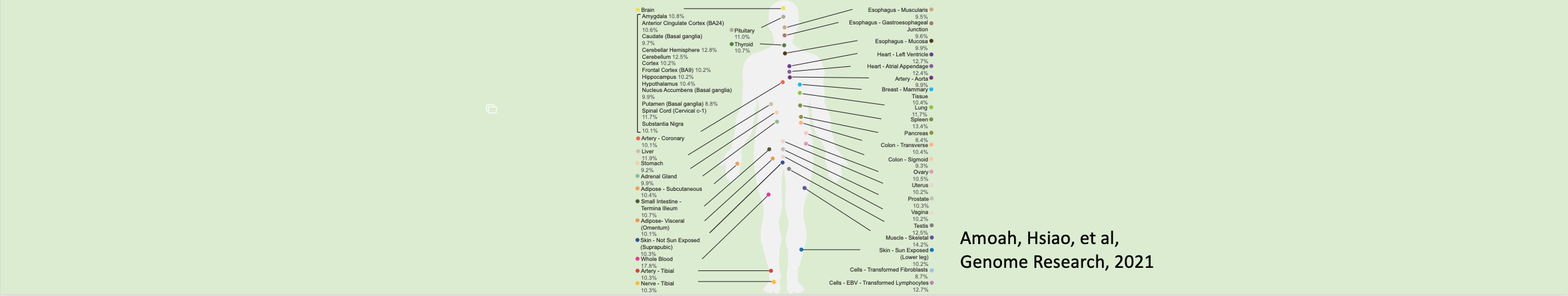
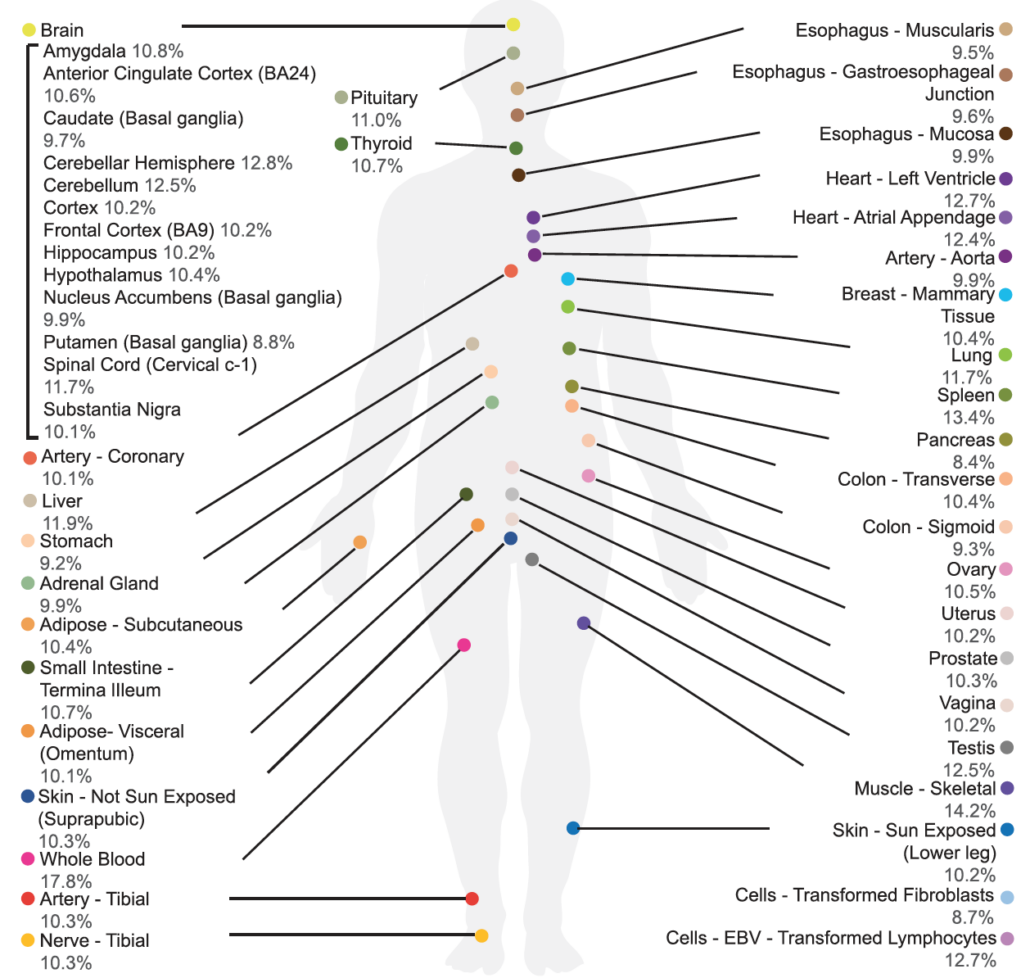
King, Alison J.; Amoah, Kofi; Zhang, Laixing; Bahn, Jae Hoon; Barney, Ryan M.; Quinones-Valdez, Giovanni; Xiao, Xinshu
Allele-specific splicing modulates protein isoforms and Alzheimer’s risk Unpublished
bioRxiv, 2026.
@unpublished{King2026,
title = {Allele-specific splicing modulates protein isoforms and Alzheimer’s risk},
author = {Alison J. King and Kofi Amoah and Laixing Zhang and Jae Hoon Bahn and Ryan M. Barney and Giovanni Quinones-Valdez and Xinshu Xiao},
url = {http://biorxiv.org/lookup/doi/10.64898/2026.02.13.705796},
doi = {10.64898/2026.02.13.705796},
year = {2026},
date = {2026-02-16},
publisher = {openRxiv},
abstract = {Abstract
Despite growing catalogs of genetic variation linked to human traits and diseases, the functional impact of most genetic variants remains poorly understood. Alternative splicing, particularly in the human brain, represents a key layer of post-transcriptional regulation that may mediate genetic effects on gene expression and protein diversity. In this study, we systematically map allele-specific alternative splicing (ASAS) events in postmortem brain tissues from the Mount Sinai Brain Bank cohort, identifying hundreds of genetically regulated splicing events across four brain regions. Using a concordance-based method, we nominate over 500 putative functional SNPs associated with ASAS, many of which overlap splicing QTLs (sQTLs), RNA-binding protein binding sites, and GWAS loci for Alzheimer’s disease (AD), brain traits, and immune phenotypes. ASAS events are enriched in genes involved in mitochondrial function and frequently occur in 5′ untranslated regions (5′ UTRs), where they are associated with protein quantitative trait loci (pQTLs), alternative start codons, and isoform-specific domain changes - highlighting an underappreciated mechanism through which noncoding variants can influence translation and proteome complexity. Importantly, we also identify a subset of ASAS events exhibiting disease-specific splicing patterns in AD brains, including functional SNPs with opposing splicing effects between AD and control groups in genes implicated in mitochondrial function and neuronal signaling. Together, our results provide a brain-specific, splicing-resolved map of regulatory variation and uncover novel mechanisms linking genetic variation to transcript and protein-level changes in AD. This work highlights the importance of allele-specific splicing analysis for interpreting noncoding variation in complex human disorders. },
howpublished = {bioRxiv},
keywords = {},
pubstate = {published},
tppubtype = {unpublished}
}
Barney, Ryan M.; Quinones-Valdez, Giovanni; King, Alison J.; Amoah, Kofi; Wang, Weijian; Xiao, Xinshu
Allele-specific alternative polyadenylation links noncoding genetic variation to Alzheimer’s disease risk Unpublished
bioRxiv, 2026.
@unpublished{Barney2026,
title = {Allele-specific alternative polyadenylation links noncoding genetic variation to Alzheimer’s disease risk},
author = {Ryan M. Barney and Giovanni Quinones-Valdez and Alison J. King and Kofi Amoah and Weijian Wang and Xinshu Xiao},
url = {http://biorxiv.org/lookup/doi/10.64898/2026.02.13.705798},
doi = {10.64898/2026.02.13.705798},
year = {2026},
date = {2026-02-15},
publisher = {openRxiv},
abstract = {Abstract
Background
Alternative polyadenylation (APA) is a crucial post-transcriptional mechanism generating isoform diversity in the nervous system. While genetic variants significantly influence gene expression, the extent to which they regulate 3’ UTR usage in the human brain remains underexplored. We aimed to characterize the landscape of allele-specific alternative polyadenylation (asAPA) and investigate its role in neurodevelopmental and neurodegenerative disorders.
Results
Analyzing 1,047 RNA-seq data from 293 Alzheimer’s disease (AD) and control donors across four brain regions, we identified 4,462 asAPA events involving 3,432 SNPs. We prioritized a core set of putative functional variants that drive consistent cis-regulatory effects across individuals. These functional SNPs are enriched for RNA-binding protein motifs, particularly those recognized by FMRP. In Fragile X Syndrome brains lacking FMRP, we observed widespread 3’ UTR shortening, with FMRP motifs enriched in the 3’ UTR extension regions of shortened transcripts, suggesting FMRP normally protects against proximal site usage. Integrating these data with population genetics, we found that asAPA SNPs significantly overlap GWAS risk loci for autism spectrum disorder (ASD), ADHD, and AD. Furthermore, comparing AD to control brains within our cohort revealed 77 asAPA genes exhibiting condition-specific shifts in allelic bias, affecting key synaptic genes including
CAMK2G
.
Conclusions
Our study uncovers a pervasive layer of cis-regulatory variation in the human brain that links noncoding genetics to transcript structure via RBP interactions. We identify FMRP as a key regulator of this process and demonstrate that asAPA provides a mechanistic bridge connecting genetic risk to neuronal pathology in both neurodevelopment and neurodegeneration.
howpublished = {bioRxiv},
keywords = {},
pubstate = {published},
tppubtype = {unpublished}
}
Analyzing 1,047 RNA-seq data from 293 Alzheimer’s disease (AD) and control donors across four brain regions, we identified 4,462 asAPA events involving 3,432 SNPs. We prioritized a core set of putative functional variants that drive consistent cis-regulatory effects across individuals. These functional SNPs are enriched for RNA-binding protein motifs, particularly those recognized by FMRP. In Fragile X Syndrome brains lacking FMRP, we observed widespread 3’ UTR shortening, with FMRP motifs enriched in the 3’ UTR extension regions of shortened transcripts, suggesting FMRP normally protects against proximal site usage. Integrating these data with population genetics, we found that asAPA SNPs significantly overlap GWAS risk loci for autism spectrum disorder (ASD), ADHD, and AD. Furthermore, comparing AD to control brains within our cohort revealed 77 asAPA genes exhibiting condition-specific shifts in allelic bias, affecting key synaptic genes including
.
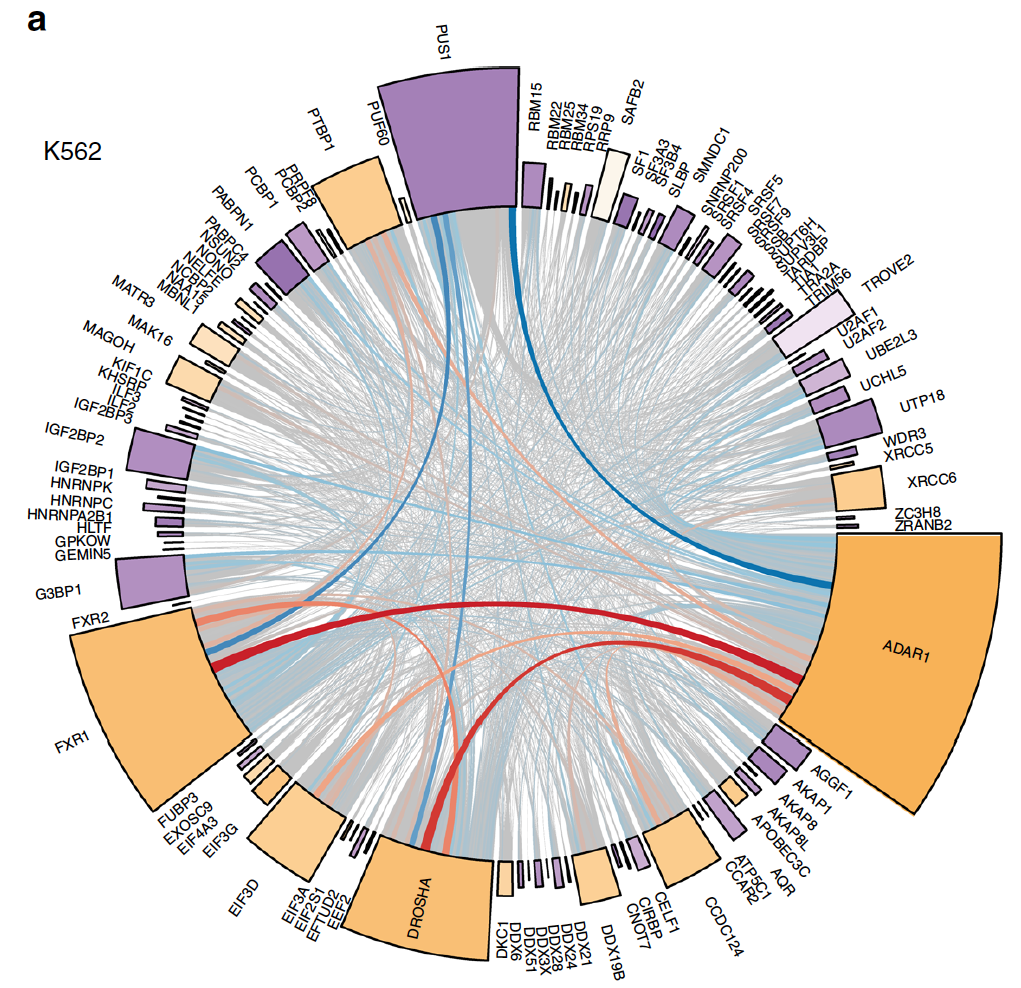
Wang, Siqi; Fraser, Anna; Xiao, Xinshu
ADAR-GPT: Learning the language of the epitranscriptome Journal Article
In: Proc Natl Acad Sci U S A, vol. 123, no. 10, pp. e2537202123, 2026, ISSN: 1091-6490.
@article{pmid41770944,
title = {ADAR-GPT: Learning the language of the epitranscriptome},
author = {Siqi Wang and Anna Fraser and Xinshu Xiao},
doi = {10.1073/pnas.2537202123},
issn = {1091-6490},
year = {2026},
date = {2026-03-01},
journal = {Proc Natl Acad Sci U S A},
volume = {123},
number = {10},
pages = {e2537202123},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2025
Seo, Hyomin; Cuddleston, Winston H.; Fu, Ting; Navarro, Elisa; Parks, Madison; Allan, Amanda; Efthymiou, Anastasia G.; Breen, Michael S.; Xiao, Xinshu; Raj, Towfique; Humphrey, Jack
Cytosine-to-uracil RNA editing is upregulated by pro-inflammatory stimulation of myeloid cells Journal Article
In: BioRxiv, 2025.
@article{nokey,
title = {Cytosine-to-uracil RNA editing is upregulated by pro-inflammatory stimulation of myeloid cells},
author = {Hyomin Seo and Winston H. Cuddleston and Ting Fu and Elisa Navarro and Madison Parks and Amanda Allan and Anastasia G. Efthymiou and Michael S. Breen and Xinshu Xiao and Towfique Raj and Jack Humphrey},
url = {https://www.biorxiv.org/content/10.1101/2025.03.14.643382v1.abstract?%3Fcollection=},
year = {2025},
date = {2025-03-17},
urldate = {2025-03-17},
journal = {BioRxiv},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Recent News
Our paper “Allele-specific splicing modulates protein isoforms and Alzheimer’s risk” is on BioRxiv. Congratulations Alison and all coauthors!
Our paper “Allele-specific alternative polyadenylation links noncoding genetic variation to Alzheimer’s disease risk” is BioRxiv. Congratulations Ryan and all coauthors!
Our paper “Genetic variation shapes human mRNA translation and disease risk” is on BioRxiv. Congratulations to Siqi!
We welcome undergraduate student Ryan Slohn to our lab!

University of California, Los Angeles 610 Charles E. Young Drive South
Terasaki Life Sciences Building, Room 2000E
Los Angeles, CA 900095-1570
Office: (310) 206-6522
Lab: (310) 206-6774
Affiliations
- Integrative Biology and Physiology
- Institute for Quantitative and Computational Biology
- Molecular Biology Institute
- Jonsson Comprehensive Cancer Center
- Bioinformatics PhD Program
- MCIP PhD Program
- Bioengineering PhD Program
- Molecular Biology PhD Program
- Graduate Programs in Bioscience
- Computational and Systems Biology Undergraduate Program