Recent Publications
2025
Seo, Hyomin; Cuddleston, Winston H.; Fu, Ting; Navarro, Elisa; Parks, Madison; Allan, Amanda; Efthymiou, Anastasia G.; Breen, Michael S.; Xiao, Xinshu; Raj, Towfique; Humphrey, Jack
Cytosine-to-uracil RNA editing is upregulated by pro-inflammatory stimulation of myeloid cells Journal Article
In: BioRxiv, 2025.
@article{nokey,
title = {Cytosine-to-uracil RNA editing is upregulated by pro-inflammatory stimulation of myeloid cells},
author = {Hyomin Seo and Winston H. Cuddleston and Ting Fu and Elisa Navarro and Madison Parks and Amanda Allan and Anastasia G. Efthymiou and Michael S. Breen and Xinshu Xiao and Towfique Raj and Jack Humphrey},
url = {https://www.biorxiv.org/content/10.1101/2025.03.14.643382v1.abstract?%3Fcollection=},
year = {2025},
date = {2025-03-17},
urldate = {2025-03-17},
journal = {BioRxiv},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Siqueira, Mirian Krystel De; Nouhi, Zaynab; Zhao, Yutian; Wang, Siqi; Xiao, Xinshu; Yang, Xia; Hulea, Laura; Villanueva, Claudio J
Protocol to perform polysome profiling in primary differentiating murine adipocytes Journal Article
In: STAR Protoc, vol. 6, no. 2, pp. 103799, 2025, ISSN: 2666-1667.
@article{pmid40315057,
title = {Protocol to perform polysome profiling in primary differentiating murine adipocytes},
author = {Mirian Krystel De Siqueira and Zaynab Nouhi and Yutian Zhao and Siqi Wang and Xinshu Xiao and Xia Yang and Laura Hulea and Claudio J Villanueva},
doi = {10.1016/j.xpro.2025.103799},
issn = {2666-1667},
year = {2025},
date = {2025-04-01},
journal = {STAR Protoc},
volume = {6},
number = {2},
pages = {103799},
abstract = {Here, we present a protocol for polysome profiling in differentiating adipocytes from the mouse stromal vascular fraction. We describe steps for lysate preparation, ultracentrifugation through sucrose gradients, mRNA isolation, RNA sequencing, and motif enrichment. This protocol enables the analysis of actively translating mRNAs and translational gene regulation by isolating polysome-bound mRNA, revealing insights into protein synthesis during adipogenesis or stimuli responses. Applications include studying translational control in adipocytes, metabolic diseases, and obesity, linking translational regulation to cellular and metabolic phenotypes. For complete details on the use and execution of this protocol, please refer to De Siqueira et al..},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Wang, Jing; Deng, Xiaoqian; Jian, Tianshen; Yin, Shanshan; Chen, Linzhi; Vergnes, Laurent; Li, Zhehao; Liu, Huoyuan; Lee, Ryan; Lim, Sin Yee; Bahn, Jae Hoon; Xiao, Xinshu; Zhu, Xianmin; Hu, Ganlu; Reue, Karen; Liu, Yizhi; Fan, Guoping
DNA methyltransferase 1 modulates mitochondrial function through bridging mC RNA methylation Journal Article
In: Mol Cell, 2025, ISSN: 1097-4164.
@article{pmid40328247,
title = {DNA methyltransferase 1 modulates mitochondrial function through bridging mC RNA methylation},
author = {Jing Wang and Xiaoqian Deng and Tianshen Jian and Shanshan Yin and Linzhi Chen and Laurent Vergnes and Zhehao Li and Huoyuan Liu and Ryan Lee and Sin Yee Lim and Jae Hoon Bahn and Xinshu Xiao and Xianmin Zhu and Ganlu Hu and Karen Reue and Yizhi Liu and Guoping Fan},
doi = {10.1016/j.molcel.2025.04.019},
issn = {1097-4164},
year = {2025},
date = {2025-04-01},
journal = {Mol Cell},
abstract = {DNA methyltransferase 1 (DNMT1) is an enzyme known for DNA methylation maintenance. Point mutations in its replication focus targeting sequence (RFTS) domain lead to late-onset neurodegeneration, such as autosomal dominant cerebellar ataxia-deafness and narcolepsy (ADCA-DN) disorder. Here, we demonstrated that DNMT1 has the capability to bind to mRNA transcripts and facilitate 5-methylcytosine (mC) RNA methylation by recruiting NOP2/Sun RNA methyltransferase 2 (NSUN2). RNA mC methylation, in turn, promotes RNA stability for those genes modulating mitochondrial function. When the DNMT1 RFTS domain is mutated in mice, it triggers aberrant DNMT1-RNA interaction and significantly elevated mC RNA methylation and RNA stability for a portion of metabolic genes. Consequently, increased levels of metabolic RNA transcripts contribute to cumulative oxidative stress, mitochondrial dysfunction, and neurological symptoms. Collectively, our results reveal a dual role of DNMT1 in regulating both DNA and RNA methylation, which further modulates mitochondrial function, shedding light on the pathogenic mechanism of DNMT1 mutation-induced neurodegeneration.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Khan, Arshad H; Bagley, Jared R; LaPierre, Nathan; Gonzalez-Figueroa, Carlos; Spencer, Tadeo C; Choudhury, Mudra; Xiao, Xinshu; Eskin, Eleazar; Jentsch, James D; Smith, Desmond J
Differing Genetics of Saline and Cocaine Self-Administration in the Hybrid Mouse Diversity Panel Journal Article
In: Genes Brain Behav, vol. 24, no. 3, pp. e70029, 2025, ISSN: 1601-183X.
@article{pmid40527326,
title = {Differing Genetics of Saline and Cocaine Self-Administration in the Hybrid Mouse Diversity Panel},
author = {Arshad H Khan and Jared R Bagley and Nathan LaPierre and Carlos Gonzalez-Figueroa and Tadeo C Spencer and Mudra Choudhury and Xinshu Xiao and Eleazar Eskin and James D Jentsch and Desmond J Smith},
doi = {10.1111/gbb.70029},
issn = {1601-183X},
year = {2025},
date = {2025-06-01},
journal = {Genes Brain Behav},
volume = {24},
number = {3},
pages = {e70029},
abstract = {To identify genes that regulate the response to the potentially addictive drug cocaine, we performed a control experiment using genome-wide association studies (GWASs) and RNA-Seq of a panel of inbred and recombinant inbred mice undergoing intravenous self-administration of saline. A linear mixed model increased statistical power for the analysis of the longitudinal behavioral data, which was acquired over 10 days. A total of 145 loci were identified for saline compared to 17 for the corresponding cocaine GWAS. Only one locus overlapped. Transcriptome-wide association studies (TWASs) using RNA-Seq data from the nucleus accumbens and medial frontal cortex identified 5031434O11Rik and Zfp60 as significant for saline self-administration. Two other genes, Myh4 and Npc1, were nominated based on proximity to loci for multiple endpoints or a cis locus regulating expression. All four genes have previously been implicated in locomotor activity, despite the absence of a strong relationship between saline taking and distance traveled in the open field. Our results indicate a distinct genetic basis for saline and cocaine self-administration, and suggest some common genes for saline self-administration and locomotor activity.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
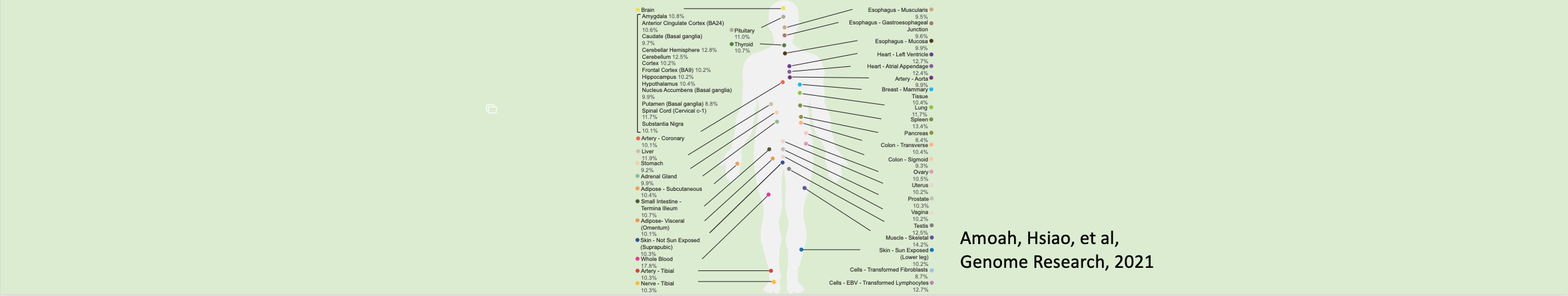
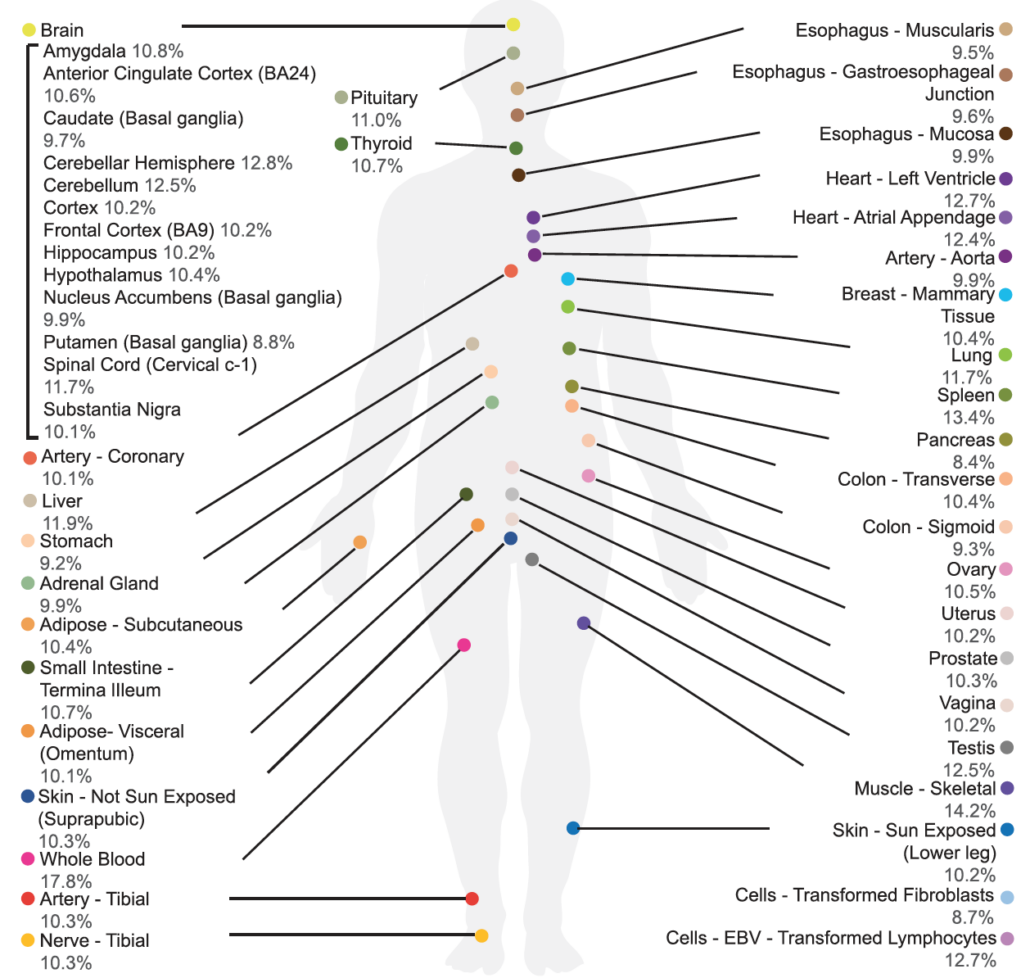
Huang, Elaine; Fu, Ting; Zhang, Ling; Yan, Guanao; Yamamoto, Ryo; Terrazas, Sari; Nguyen, Thuy Linh; Gonzalez-Figueroa, Carlos; Khanbabaei, Armen; Bahn, Jae Hoon; Varada, Rajagopal; Amoah, Kofi; Hervoso, Jonatan; Paulsen, Michelle T; Magnuson, Brian; Ljungman, Mats; Li, Jingyi Jessica; Xiao, Xinshu
Genetic variants affecting RNA stability influence complex traits and disease risk Journal Article
In: Nat Genet, 2025, ISSN: 1546-1718.
@article{pmid40913182,
title = {Genetic variants affecting RNA stability influence complex traits and disease risk},
author = {Elaine Huang and Ting Fu and Ling Zhang and Guanao Yan and Ryo Yamamoto and Sari Terrazas and Thuy Linh Nguyen and Carlos Gonzalez-Figueroa and Armen Khanbabaei and Jae Hoon Bahn and Rajagopal Varada and Kofi Amoah and Jonatan Hervoso and Michelle T Paulsen and Brian Magnuson and Mats Ljungman and Jingyi Jessica Li and Xinshu Xiao},
doi = {10.1038/s41588-025-02326-8},
issn = {1546-1718},
year = {2025},
date = {2025-09-01},
journal = {Nat Genet},
abstract = {Gene expression is modulated jointly by transcriptional regulation and messenger RNA stability, yet the latter is often overlooked in studies on genetic variants. Here, leveraging metabolic labeling data (Bru/BruChase-seq) and a new computational pipeline, RNAtracker, we categorize genes as allele-specific RNA stability (asRS) or allele-specific RNA transcription events. We identify more than 5,000 asRS variants among 665 genes across a panel of 11 human cell lines. These variants directly overlap conserved microRNA target regions and allele-specific RNA-binding protein sites, illuminating mechanisms through which stability is mediated. Furthermore, we identified causal asRS variants using a massively parallel screen (MapUTR) for variants that affect post-transcriptional mRNA abundance, as well as through CRISPR prime editing approaches. Notably, asRS genes were enriched significantly among a multitude of immune-related pathways and contribute to the risk of several immune system diseases. This work highlights RNA stability as a critical, yet understudied mechanism linking genetic variation and disease.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Recent News
Carlos, Jonatan and Ryo defended their dissertations! Congratulations, Dr. Gonzalez-Figueroa, Dr. Hervoso and Dr. Yamamoto!!!
Congratulations to Angela for being awarded the MARC scholarship!
Ryo’s paper on “Cell-Type-Resolved Isoform Atlas of Human Tissues Reveals Age and Alzheimer’s Disease-Associated Splicing Changes” is on BioRxiv! Congratulations, Ryo and coauthors!
Our paper “Long-read RNA-seq demarcates cis- and trans-directed alternative RNA splicing” is published at Nature Communications. Congratulations Gio and Kofi!
We are glad to have contributed to this paper: Meta-analysis of genetic regulation of RNA editing in the human brain identifies new genes underlying neurological disease. A great collaboration with Winston, Jack, Towfique and others on the team!

University of California, Los Angeles 610 Charles E. Young Drive South
Terasaki Life Sciences Building, Room 2000E
Los Angeles, CA 900095-1570
Office: (310) 206-6522
Lab: (310) 206-6774
Affiliations
- Integrative Biology and Physiology
- Institute for Quantitative and Computational Biology
- Molecular Biology Institute
- Jonsson Comprehensive Cancer Center
- Bioinformatics PhD Program
- MCIP PhD Program
- Bioengineering PhD Program
- Molecular Biology PhD Program
- Graduate Programs in Bioscience
- Computational and Systems Biology Undergraduate Program